Working with reverse strand proteins requires special attention, as the position arithmetic and logic needs to be worked out with care. As developers, we need to have a documented procedure to be able to check amino acid and nucleotide sequences and this post is an attempt in that direction.
First, given an accession table and mascot results (in DAT) format, the following bash command outputs proteins of interest in the mascot search
1
| |
We get lines as the followings:
1 2 3 4 5 6 7 8 9 10 11 12 13 | |
Let’s pick a protein match there ARB1_YEAST. We search for that protein in UniProtKB and we find it as shown in image below
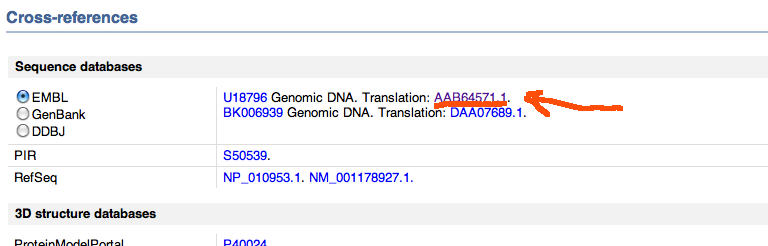
We see there the locus name YER036C, which we would find in the accession.txt if we looked for our protein of interest. In the UniProtKB there is a cross-refs link which we click. It just points to one of the sections at the bottom of the page as shown here
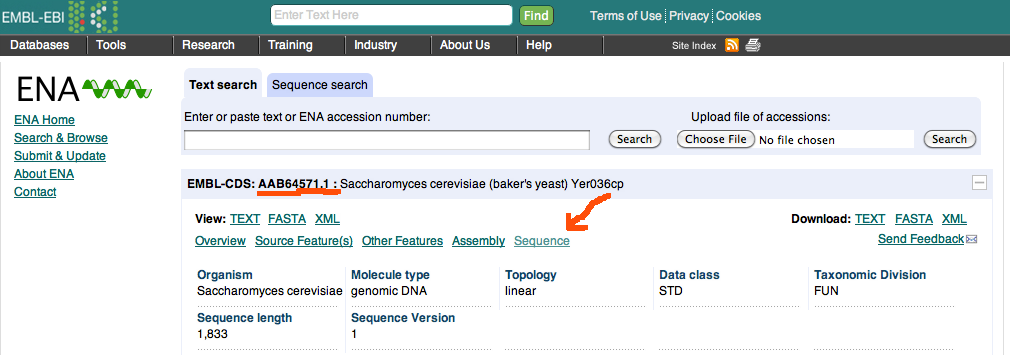
We follow the link to the EMBL sequence database, particularly the translation code shown. Follow that in new window as it is another website, EMBL-EBI, particularly the page describing the sequence for that protein
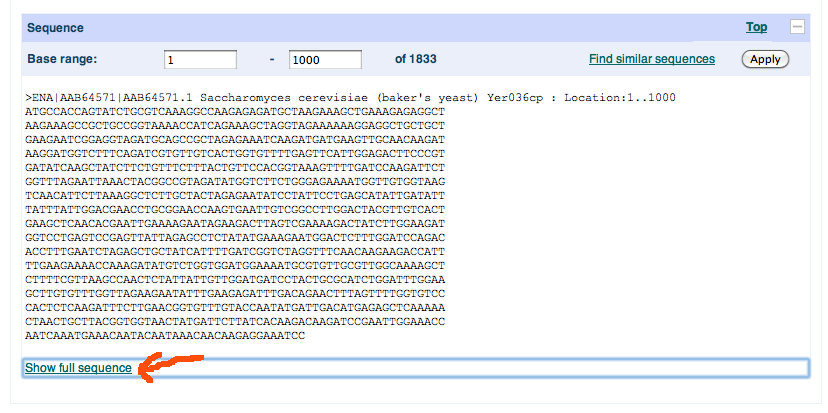
as the preview above should confirm. There we just go to sequence (see red arrow), a link to the bottom, where we see the sequence as displayed in here
So, just click the “Show full sequence” link (JavaScript) to reveal the full sequence if it is too long. Voila, the amino acid sequence!
Bear in mind that this whole exercise assumes a reference genome. You can see it just below the locus name in the first screenshot above. Hope this instructions help to navigate the complex web of proteomic/genomic data.